We offer the plant research community and companies access to our latest interactomics tools, covering AP-MS and PL-MS (Turbo-ID) as complementary approaches. We also offer service for XL-MS based on tandem affinity purification of stable protein complexes.
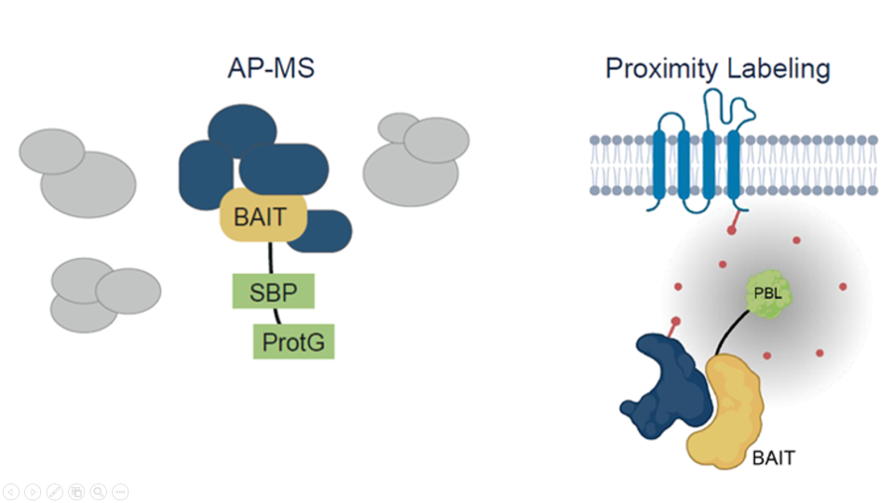
Experiments are performed under Research Service Agreement, following well-established protocols mainly developed in Arabidopsis PSB-D cell suspension culture. As starting material, either cell cultures, callus tissue, seedlings or isolated plant tissues can be used. We have extensive experience with interactomics analyses in Arabidopsis. Moreover, our protocols have been successfully transferred for analyses in maize, rice, and canola. For efficient AP-MS, we prefer to use Protein A/G-based tags. Alternatively, also AP-MS based on GFP or other tags can be requested. For proximity labeling, we follow the Turbo-ID approach. We have also ample experience with tandem affinity purification for the isolation of stable protein complexes at high purity. The latter we can apply in cross linking experiments that deliver structural analysis of the protein complex based on the identification of cross linked peptides.
For identification of isolated proteins, we have access to state of the art mass spectrometry instruments (e.g. Orbitrap Fusion Lumos and Q-Exactive HF). In order to pinpoint specific interactors, non-specific proteins are filtered out using a multitude of methods, integrating label-free quantitative MS tools and covering either a large dataset approach, control experiments or conservation of non-specific background proteins across plant species.
For cell cultures, we start from delivered bait clones. For experiments in plants, we start from delivered plant tissue. More details and conditions are provided after consulting us at [email protected]